Responding to Covid-19
Across the University experts have come together to focus their talent and energies on tackling the pandemic.
On 18 March, the Vice-Chancellor, Professor Stephen J Toope, announced that the University was moving into the red phase response to the Covid-19 pandemic. “We will need all the fortitude, resilience and generosity of our community to get us through,” he said. “I am confident that this crisis will bring out the best in all of us.” Across the University, world-leading experts from immunology to engineering have proved him right, as they focus their energies and talents on tackling the Covid-19 pandemic.
The response was led by the Cambridge Institute of Therapeutic Immunology and Infectious Disease (CITIID), one of the University’s newest institutes – it opened at the end of last year.
As Director of the Cambridge Institute of Therapeutic Immunology and Infectious Disease, Professor Smith has had an intense few months – but the team’s work is already having an impact on the national and international response to Covid-19. Eye of Science/Science Photo Library
“When it was clear that there was going to be a lockdown, we decided to pivot the entire Institute to focus on Covid-19 research,” says CITIID’s Director, Professor Ken Smith. “All the groups in CITIID work at the interface between pathogens and the immune system, and indeed the Institute was designed to respond to pandemics. But what we’ve achieved within weeks would have taken over a year in a normal situation. And that’s down to incredible collaboration with Addenbrooke’s [part of the Cambridge University Hospitals NHS Foundation Trust] plus the Cambridge Biomedical Research Centre. And we have been humbled by unflagging support throughout the pandemic from volunteers, from across the University, whether scientists helping with laboratory research or couriering blood samples or senior technicians volunteering as cleaners.”
We pivoted the entire Institute to focus on Covid-19. The Institute was designed to respond to pandemics, but we’ve achieved in weeks what would normally take over a year
Professor Ken Smith
CITIID is home to the UK’s largest academic microbiological Containment Level 3 facility and 150 scientists and clinicians. It works with a range of collaborators from across the UK and beyond. It is integrated with the NHS both locally, through Addenbrooke’s and the Royal Papworth Hospital, and, nationally, in particular through the National Institute for Health Research (NIHR).
The team knew the initial response had to focus on staff on the frontline, and that testing both patients and healthcare workers would be key. Medical researcher Dr Helen Lee developed the technology behind the SAMBA II machine while working at the haematology department in Cambridge. Originally intended for HIV testing in Africa, her spinout company, Diagnostics for the Real World, rapidly adapted it for Covid-19. Meanwhile, Professor Ravi Gupta of CITIID led the first validation study. The machine is now being used extensively at Addenbrooke’s, and more widely in the NHS. “It gives a result in minutes to an hour, which means you can make rapid decisions,” says Smith. “This allows patients to be sent quickly to the appropriate wards, speeding up medical care and reducing the chance of virus transmission. That has been a game-changer.”
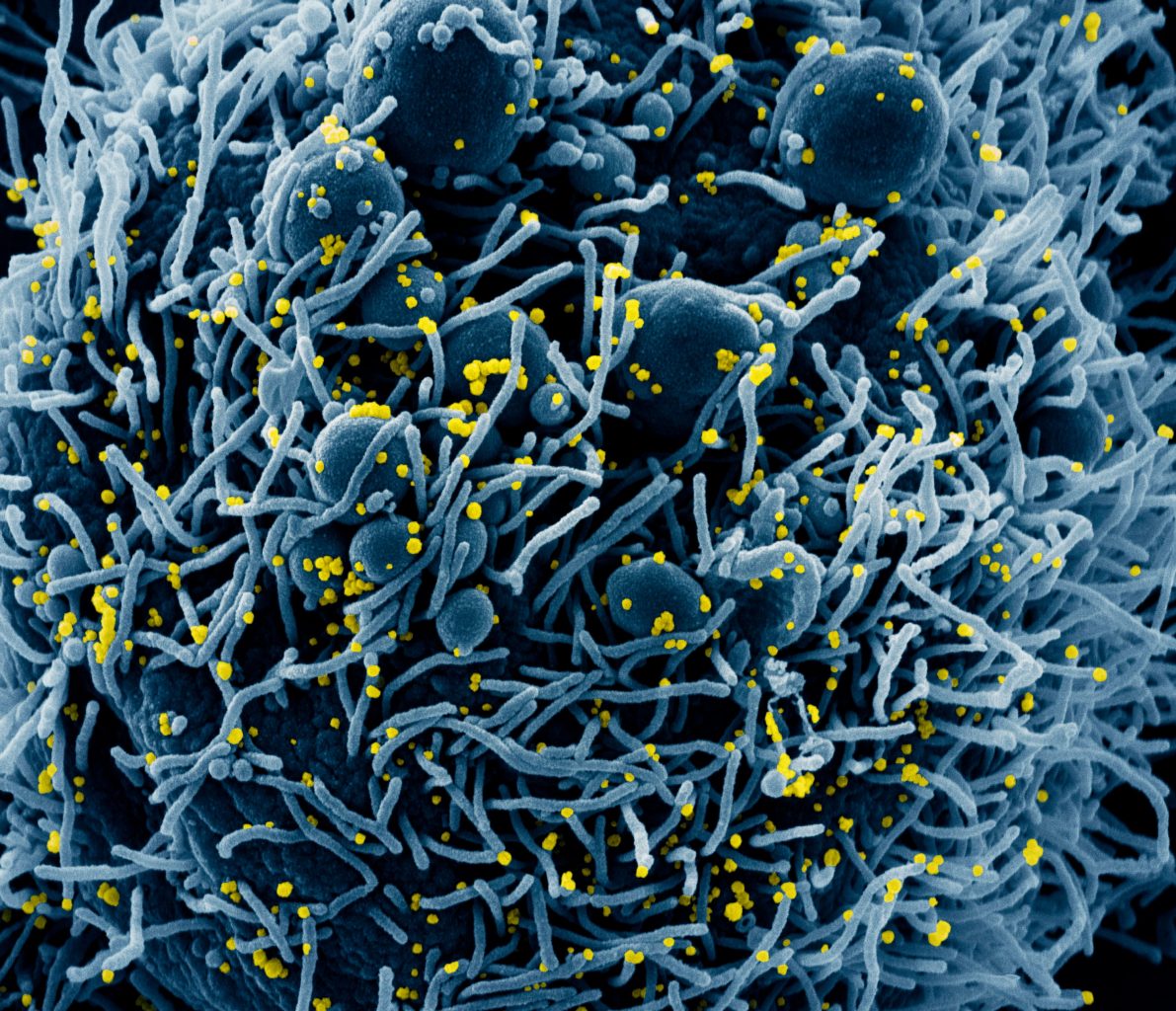
Coloured scanning electron micrograph of Sars-CoV-2 coronavirus particles (yellow) on an apoptotic cell (blue) from a US patient sample. Sars-CoV-2 is an enveloped RNA (ribonucleic acid) virus that causes respiratory infection that can lead to fatal pneumonia. NIAD/National Institutes of Health/Science Photo Library
Another approach is a polymerase chain reaction (PCR) test, modified by Dr Martin Curran. This test allows scientists to extract a minuscule amount of RNA from the virus and copy it millions of times, creating an amount large enough to confirm presence of the virus. However, because the virus itself is infectious, samples have to be carefully processed. Understandably, the safety requirements slow down the testing process. In response, Professor Stephen Baker has been working on augmenting both capacity and speed of testing. His team at CITIID have used a method of inactivating the virus at the point of sampling. This means that tests can now be processed in Containment Level 2 facilities, which are more widely available and have fewer restrictions on their use. Their method is based on previous work led by Professor Ian Goodfellow and colleagues in the Department of Pathology. From initial nasal swab to result, the entire process takes just four hours. This enabled the team, working with academic infectious diseases physician Dr Michael Weekes, to begin screening healthcare workers at Addenbrooke’s Hospital for Sars-CoV-2. Initially three per cent of working hospital staff tested positive, but after six weeks of screening this has been reduced to fewer than one in 1,000.
The team is developing an open-source artificial intelligence tool that combines chest imaging data with clinical data to support the diagnosis and triage of Covid-19 in the UK
Professor Carola-Bibiane Schönlieb
The University, in partnership with AstraZeneca and GSK, has also risen to the UK government’s challenge to provide sustainable testing capacity for Covid-19. A new state-of-the-art laboratory has been set up in a matter of weeks at the University’s Anne McLaren Building, which uses advanced automation to deliver tests at rapid scale and pace. It was an enormous undertaking that involved everything from recruiting and training staff to bringing in equipment and setting up scientific protocols. At maximum capacity, the lab can run tens of thousands of tests a day to contribute to the national effort to tackle Covid-19.
Testing for antibodies, to see if people have had the virus, will also be crucial to future efforts to control the disease. At CITIID, teams are assessing commercially available serology tests alongside ones produced by the Institute and its collaborators. These will be moving in-clinic “very soon”, says Smith. “These tests are important in that they tell us who has been exposed, and potentially whether you have protective immunity or not. We know we can measure antibodies in the blood. What we don’t know yet is whether or not they kill the virus and provide immunity, or how long this protection will last. All the serum that we and others are collecting around the country is being used to try to answer those questions.”
Patient trials enabling researchers to learn more about the disease and possible therapies are now up and running. While most people recover from Covid-19, a proportion experience an unusually aggressive inflammatory response. This drives damage to the lungs and other organs, which can be fatal. If we understand more about why some have this response and some don’t, we could use targeted therapies to reduce inflammation more effectively, says Smith. “So we have started recruiting patients with different degrees of severity of Covid-19, from asymptomatic patients all the way through to patients in intensive care and on ventilators. We have recruited 250 patients and are now doing immune analyses to compare the stages of disease.” The Institute’s Professor David Jayne has also set up a national trial currently under way to compare the effects of two drugs, Ravulizumab and Baricitinib.

Applied mathematician Professor Schönlieb’s project will process and analyse complex, multi-stream patient data such as imaging, clinical information and laboratory results. The team has already begun receiving large imaging and clinical datasets from Austria, China, Italy and the UK. NIAD/National Institutes of Health/Science Photo Library
This is a cross- and multi-disciplinary response. Carola-Bibiane Schönlieb, Professor of Applied Mathematics and Head of the Cambridge Image Analysis (CIA) group, and Evis Sala, Professor of Oncological Imaging, are leading a team developing an open-source artificial intelligence tool that combines chest imaging data and clinical data to support diagnosis and triage of Covid-19 in the UK. The project will process and analyse complex, multi-stream patient data such as imaging, clinical information and laboratory results. The team has already begun receiving large imaging and clinical datasets from Austria, China, Italy and the UK.
AI is also being used to help decision-making at a hospital-wide level. Using anonymised data from Public Health England, Mihaela van der Schaar, the John Humphrey Plummer Professor of Machine Learning, Artificial Intelligence and Medicine, has built a working proof of concept that demonstrates the potential use of machine learning in helping to manage scarce resources like ICU beds and ventilators – although, she emphasises, the decisions will still be made by healthcare professionals on the basis of their organisation’s priorities and policies.
“Among other things, we wanted to demonstrate that machine learning techniques can accurately predict how Covid-19 will impact resource needs – such as ventilators and ICU beds – at a patient and a hospital level, thereby giving a reliable picture of future resource usage and enabling healthcare professionals to make well-informed decisions about how these scarce resources can be used to achieve the maximum benefit,” she says. “Based on the data we received from Public Health England, we now have a proof-of-concept demonstrator showing that this can be done, in the form of a new system we call Adjutorium. Once trained with patient data, Adjutorium was able to make highly accurate predictions about the patients whose data we used for verification.”

In 2018, Mathematician Professor Julia Gog worked with the BBC to conduct the UK’s largest citizen science experiment, using location data from mobile phones to map how pandemic influenza might spread. Now, using that data, she has been mapping the spread of Covid-19 to work out how to reduce the number of people one infected person goes on to infect. NIAD/National Institutes of Health/Science Photo Library
Of course, Covid-19 is just the latest in a string of pandemics: A(H1N1) influenza being the most recent prior to the current one. Repurposing knowledge from past pandemics has been crucial to understanding how the novel coronavirus behaves. In 2018 Julia Gog, Professor of Mathematical Biology (DAMTP), and her team were behind the UK’s largest citizen science experiment in collaboration with the BBC, using location data from mobile phones to map how pandemic influenza might spread across the UK. Now, she and other members of her research group have been modelling and mapping the spread of Covid-19, using knowledge gleaned from the BBC project to work out how to reduce the number of people an infected person infects – the reproductive ratio, or R.
“Public health experts have been saying for decades that when it comes to pandemic flu, it wasn’t a matter of ‘if’, it was a matter of ‘when’,” says Gog. “And now that this coronavirus pandemic is here and things are changing every day, we’ve got to get information out there quickly, but make sure that it’s useful information that can help inform good policy.”
Things are changing every day. We’ve got to get our information out there quickly, but still make sure that it’s useful and can help inform good policy
Professor Julia Gog
Understanding the mechanics of the virus will be crucial to finding effective treatments. Like other dangerous viruses such as Zika, Ebola and influenza, the Covid-19 virus’s genome is made of RNA (rather than DNA), which means it has a higher rate of mutation. However, counterintuitively, this could work against the success of the virus, rather than for it.
“Dependencies that arise from unusual biology may serve as chinks in the virus’s armour,” says Professor Gerard Evan, Head of the Department of Biochemistry. Several teams in the Department of Biochemistry are studying exactly what those chinks might be: Professor Ben Luisi’s group, for example, is targeting a specific structured RNA element in the Covid-19 genome that is also found in Sars, Mers and other coronaviruses. It’s not known how this element affects the virus lifecycle – but the fact that it’s present in all these viruses suggests it plays a fundamental role.
The University is also taking a leading role in the Covid-19 Genomics UK (COG-UK) Consortium, led by Sharon Peacock, who is Professor of Public Health and Microbiology at the University and Director of the National Infection Service for Public Health England. Comprising the NHS, public health agencies, the Wellcome Sanger Institute and numerous academic institutions, the £20m project enables large-scale, rapid sequencing of the disease and the sharing of intelligence with hospitals, regional NHS centres and the government.
You look at the diversity in the genome and try to wind the clock back: what are the mutations, when did they occur? Where did this strain emerge and how does it fit into the UK pattern?
Dr Charlotte Houldcroft
Samples from patients with confirmed cases of Covid-19 will be sent to a network of sequencing centres across the UK. The University, together with the Wellcome Sanger Institute, will co-ordinate the collaboration between expert groups to analyse the genetic code of Covid-19 samples circulating in the UK. That will give public health agencies and clinicians a unique tool to combat the virus.
By looking at the whole virus genome in people who have had confirmed Covid-19, scientists can monitor changes in the virus at a national scale to understand how the virus is spreading and whether different strains are emerging. This will help clinical care of patients and save lives. “This virus is one of the biggest threats our nation has faced in recent times and crucial to helping us fight it is understanding how it is spreading,” says Peacock. “Harnessing innovative genome technologies will help us tease apart the complex picture of coronavirus spread in the UK, and rapidly evaluate ways to reduce the impact of this disease on our society.”
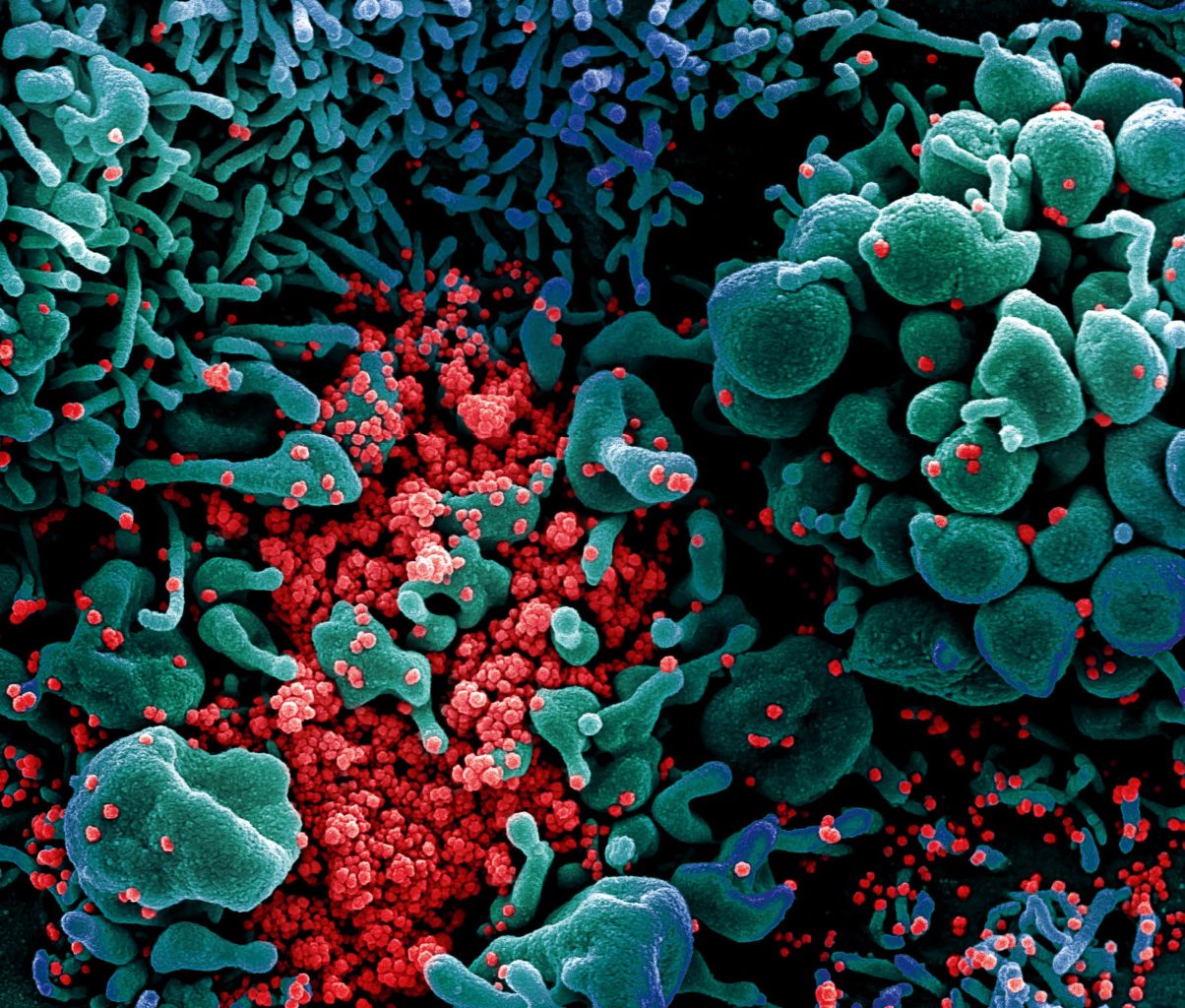
This coloured scanning electron micrograph shows Sars-CoV-2 coronavirus particles (red) emerging from a heavily infected apoptotic cell (blue) from a US patient sample. NIAD/National Institutes of Health/Science Photo Library
Virus evolution researcher Dr Charlotte Houldcroft is on the frontline of the sequencing effort. She works in a tent inside a lab on the Cambridge Biomedical Campus as part of a team led by Professor Ian Goodfellow. The lab can sequence the genomes of between 24 and 70 virus samples a day.
“You look at the diversity in the genome and try to wind the clock back: working out what are mutations, when they occurred, and so where this strain emerged and how it fits into the UK pattern,” says Houldcroft. “Some of the first samples of the virus in the UK – the Chinese visitors in York and the cluster in Brighton – are not related that closely to anything we’re seeing in the country now. It suggests the tracking and tracing at the beginning worked. It was not until it started to spread across Europe and the US that we get multiple introductions across the UK. Virus clusters in Wales, for example, had contributions from all over the world. There’s not a single Welsh strain.”
At the Department of Engineering, engineers are using modelling tools originally designed to improve the efficiency of factories to help Addenbrooke’s Hospital manage the Covid-19 emergency. “It’s looking at the physical flow of patients and projecting admissions rates into the future – identifying where ‘bottlenecks’ might occur, and where the hospital might need to scale up beds, ventilators, oxygen and staff as part of their Covid-19-orientated activities,” says Dr Ajith Parlikad at the Institute for Manufacturing.
Modelling will also help the hospital better understand staffing levels during disruptions such as the Covid-19 outbreak. “With close input from the hospital, we’ve been able to pick from a series of industrial techniques and apply the most useful ones to this new setting,” says Duncan McFarlane, Professor of Industrial Information Engineering. “Instead of production lines, we’re now looking at hospital wards, and, rather than products or raw materials, we are examining the flow of patients and supplies.”
But while understanding Covid-19 and all its ramifications is just beginning, Smith is already looking to the future. “Our long-term strategy cannot be anti-inflammatories for severe disease because you have to stop it getting to that stage. It’s either going to be vaccines or anti-viral drugs. We have put a lot of resource into seeking these longer-term solutions.
“There are hundreds of known coronaviruses, and Sars-CoV-2 is the third new one to cross into humans in less than 20 years. So this is not going to stop – these things will keep coming. And we are planning to come out of our work on this pandemic with a much better foundation in place to support a rapid research response to the next one.”
Read more about the University’s response to the COVID-19 pandemic.